Email: info@chuchangbiotech.com Tel: 86-15623560702
Email: info@chuchangbiotech.com Tel: 86-15623560702
1. KRAS gene and related pathways
1) The concept of KRAS gene
The full name of the KRAS gene is Kirstenratsarcomaviraloncogenehomolog, which translated into Chinese is "Kirsten rat sarcoma virus oncogene homolog". The protein encoded by the KARS gene is a small GTPase (smallGTPase), which belongs to the RAS superprotein family.
2) Classification of KRAS genes
In the human genome, there are 2 KRAS genes. One is KRAS1, located on the short arm of chromosome 6; the other is KRAS2, located on the short arm of chromosome 12. Among them, KRAS1 is a "pseudogene" and cannot be transcribed into RNA, so it has no function. KRAS2 is the "true gene" that can be transcribed and translated into protein and has biological activity.
Note: Usually, the KRAS gene and protein studied in companies and literature reports refer to the "KRAS2" gene and its protein product.
The KRAS gene belongs to the RAS gene family. Among the RAS gene family, there are NRAS (neuroblastoma-RAS) and HRAS (Harvey-RAS). HRAS, KRAS and NRAS are located on chromosomes 11, 12 and 1 respectively, and are all involved in intracellular signal transmission.
There are 2 variants of the KRAS protein. These two variants are due to the fact that exon 4 has two different splicing methods during the splicing process of RNA, resulting in two types of mRNA, which are then translated into two types of proteins (variants). These two proteins are called "KRAS4A" and "KRAS4B" respectively. The expression amount of KRAS4B is significantly higher than that of KRAS4A. Generally, KRAS4B is 5 times that of KRAS4A.
3) Structure and location of KRAS protein
KRAS protein has 188 amino acids and its molecular weight is 21.6KD. A guanine-binding protein with GTPase activity.
The KRAS protein is located on the inside of the cell membrane and is connected to the cell membrane through a farnesyl modification group [1]. Farnesoyl is added to the KRAS protein through post-translational protein modification under the action of farnesyl transferase [2] [3].
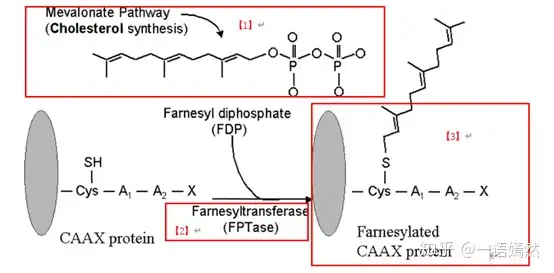
source:Farnesyltransferaseinhibitorsinhematologicmalignancies:newhorizonsintherapy
upstream signaling pathways
In normal cells, receptor monomers such as EGFR, HER2, ErbB3 and ErbB4 on the cell membrane combine with ligands outside the cell membrane to form dimers, which autophosphorylate and then phosphorylate downstream signaling proteins. One of the signaling pathways activates Grb2-Shc, which in turn activates the SOS protein, which in turn activates the KRAS protein.
KRAS pathway
Within the cell, the KRAS protein transitions between inactive and activated states. KRAS is in the inactive state when it binds to guanosine diphosphate (GDP), and it is in the inactive state when it binds to guanosine triphosphate (GTP). , it is active and can activate downstream signaling pathways.
downstream signaling pathways
KRAS is in an inactive state in most cells. When it is activated, it can activate multiple downstream signaling pathways, including the MAPK signaling pathway, PI3K signaling pathway, and Ral-GEFs signaling pathway. These signaling pathways play important roles in promoting cell survival, proliferation, and cytokine release.
One type is guanine nucleotide exchange factors (GEFs), which catalyze the binding of KRAS to GTP, thus promoting the activation of KRAS, including SOS proteins (GEFs/guanosine releasing factors/guanylate exchange factors). The other type is GTPase (GTPase) activating proteins (GAPs), which can promote the hydrolysis of GTP bound to KRAS into a GDP-terminated active state, thus inhibiting the activity of KRAS.
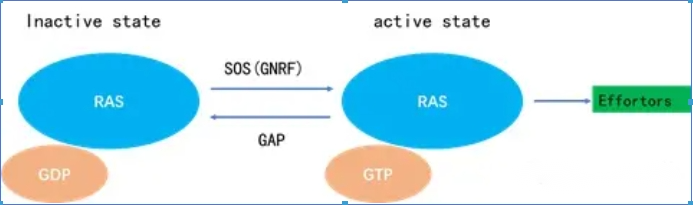
a) RAS is a binary molecular switch controlled by the GDP/GTP cycle: when the KRAS protein is not activated, it is tightly bound to GDP and is in an "off" state; after receiving the signal, guanylate exchange factors GEFs (SOS) are recruited Binds to RAS on the cell membrane and releases GDP. After KRAS is activated by SOS protein, it turns to bind to GTP.
b) The KRAS protein that binds to GTP has phosphokinase activity and represents the "on" state, which can further activate downstream proteins. Note: When there is no stimulus signal, this on-off state transition is very slow.
c) Activated downstream signaling proteins: Raf protein family, PI3K protein, PLC-ε, RALGDS, TIAM1, RIN1 and other downstream proteins [1].
d) After the downstream signaling pathways are opened, the downstream signaling pathways are further activated [2], thereby initiating cell proliferation, cell migration and other cell functions [3].
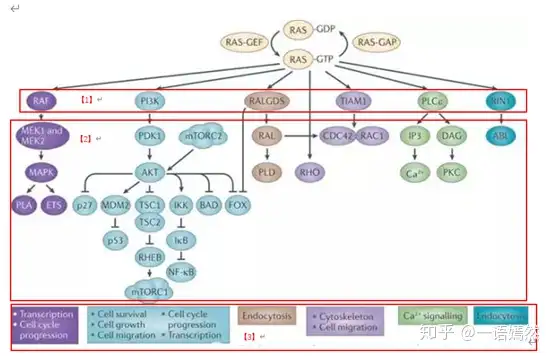
source:NatureReviews|Cancer
2. Mutations in the KRAS oncogene
Because RAS are central to the axis of many important cell signaling networks, they have been implicated in many cancer hallmarks. RAS proteins are so named because they were first identified in studies of rat sarcoma caused by a potent oncogenic virus. RAS is the most commonly mutated oncogene in human cancers, and RAS protein activation caused by mutations has been found in approximately 1/5 of all human tumors.
KRAS is the most common subtype in the RAS family, and KRAS gene mutations account for 85% of the total RAS gene mutations (NRAS (12%) followed by HRAS (3%)). In human cancers, KRAS gene mutations occur in nearly 90% of pancreatic cancers, 30-40% of colon cancers, 17% of endometrial cancers, and 15-20% of lung cancers (mostly NSCLC). It also occurs in cancer types such as cholangiocarcinoma, cervical cancer, bladder cancer, liver cancer, and breast cancer. In other words, there is a high proportion of KRAS gene mutations in the various cancers mentioned above.
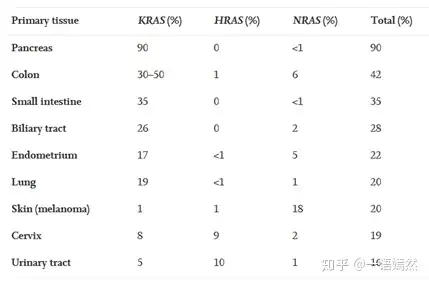
Data Sources:Liuetal.,(2019).TargetingtheuntargetableKRASincancertherapy.ActaPharmaceuticaSinicaB,https://doi.org/10.1016/j.apsb.2019.03.002
Among KRAS gene mutations, 97% are mutations in the 12th or 13th amino acid residue. The most important of these are the three mutations G12D, G12V, and G13D. Structural studies show that most of these gene mutations interfere with the ability of KRAS to hydrolyze GTP.
Three major mutations in the KRAS gene

KRAS binds very strongly to GDP or GTP, and its (KRAS) binding affinity coefficient to GTP is at the PicoMolar level. When KRAS binds to GDP, it has no kinase activity; when KRAS binds to GTP, it has kinase activity. Whether wild-type KRAS binds to GDP or GTP is regulated by upstream signals.
When G12D, G12V, and G13D mutations occur in KRAS, by destroying GAP activity, KRAS will remain bound to GTP, locking KRAS in a tyrosine kinase active state, and continuously activate downstream signaling pathways (such as PI3K, RAF-MEK-ERK (MAPK), RAL-GEF, etc.). Once these downstream signaling pathways are turned on, they will stimulate cell proliferation and migration, and ultimately promote tumorigenesis.
Let us use an analogy to illustrate the consequences of this mutation: KRAS is compared to the keeper of a bank vault (cell proliferation, migration ability). Under normal circumstances, the custodian (KRAS) can open the vault (cell proliferation, migration) only if the bank president (upstream signaling protein) comes with the key (activation signal). Now that the treasury keeper (KRAS) has stolen the key to the treasury and opened the door to the treasury, the treasury (cell proliferation and migration) is out of control.
RAS plays a pivotal role in the signal regulation of cell growth. Upstream cell surface receptors such as EGFR (ErbB1), HER2 (ErbB2), ErbB3, and ErbB4 [1], after receiving external signals [2], will transmit the signal to the downstream [4] through RAS protein [3]. This in turn stimulates cell proliferation and migration.

Source: Chen Weixue Gene
KRAS is an important member of the RAS protein. Mutations in the KRAS gene will directly affect the efficacy of anti-tumor drugs targeting the EGFR gene. At the same time, whether the KRAS gene is mutated is also an important indicator of tumor prognosis.
If a cancer patient has an activating mutation of KRAS in his tumor, then EGFR-targeted therapeutic drugs, such as cetuximab, Iressa, etc., are usually not very effective. Because KRAS bypasses the inhibitory effect of drugs on EGFR. In addition, the prognosis of tumor patients with KRAS activating mutations is generally not very good, and the survival period is often significantly shorter than that of patients without KRAS activating mutations.
Therefore, whether there are activating mutations in the KRAS gene in tumors is very important for predicting whether EGFR inhibitor drugs (treatments) will be effective. For this reason, the detection of activating mutations in the KRAS gene is now one of the important companion diagnostics for tumor-targeted drugs.
Studies have shown that in addition to directly promoting the proliferation and survival of tumor cells, oncogenic KRAS gene mutations can also affect the tumor microenvironment.Oncogenic KRAS induction affects cells surrounding the stroma (such as fibroblasts, innate and adaptive immune cells) in a paracrine manner, and these stromal cells in turn promote cancer malignancy.
Tumor cells carrying KRAS gene mutations can secrete a variety of cytokines, chemokines, and growth factors, including IL-6, IL-8, IL-23, CCL9, hedgehog, etc. These factors can reprogram stromal cells in the tumor microenvironment. For example, IL-6 and IL-8 maintain the stromal inflammatory phenotype in pancreatic and lung cancers. Granulocyte-macrophage colony-stimulating factor (GM-CSF) secreted by tumor cells can stimulate the infiltration of myeloid-derived suppressor cells (MDSC) into tumors, thereby inhibiting the anti-tumor immune response.
The impact of KRAS mutations on the tumor microenvironment
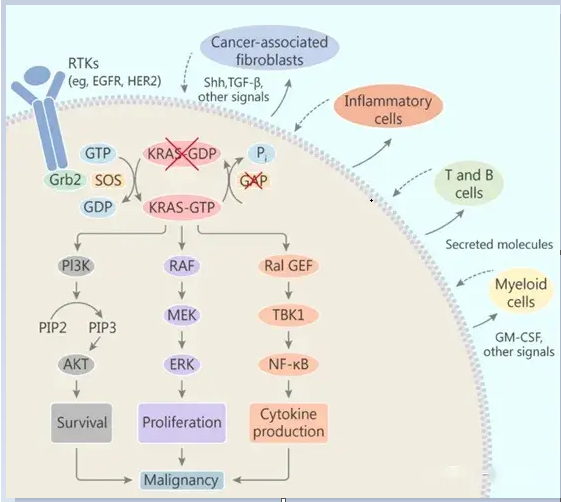
Data Sources:
Liuetal.,(2019).TargetingtheuntargetableKRASincancertherapy.ActaPharmaceuticaSinicaB,https://doi.org/10.1016/j.apsb.
Contact: Mr.Ren
Phone: 15623560702
Tel: 86-15623560702
Email: info@chuchangbiotech.com
Add: Chuangye Street, Hongshan District,Wuhan,Hubei Province,China.
We chat
